A software platform to integrate, analyze, and visualize genomic data within a systems biology context
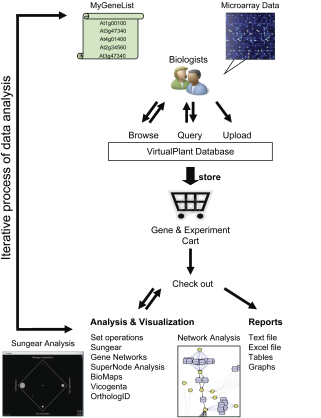 The ultimate goal of our Plant Systems Biology studies is to predictively model how internal and external perturbations affect processes, pathways, and networks controlling plant growth and development [1]. Succeeding in this endeavor will allow us to (i) explain mechanistically how molecular network changes occur to evoke responses and (ii) predict molecular network states under untested conditions or after modifications. Such predictive network models are useful for developing strategies that intervene in molecular networks for practical applications. To enable such Systems Biology approaches in plants, we developed informatic, visualization, and statistic tools to enable dynamic modeling and visualization of molecular networks operating within cells. The Arabidopsis multinetwork is a first step towards a molecular wiring diagram of the plant cell. This qualitative network model of the Arabidopsis genome contains multiple sources of evidence for all available information on gene, protein, and RNA interactions, including 16,562 nodes and 97,423 interactions [1]. Queries of this qualitative Arabidopsis multinetwork with quantitative transcriptome data result in sub-networks that can be analyzed for network properties, including connectivity, to provide focus for biological hypotheses and testing. Examples of validated hypotheses derived from queries of this multinetwork are described in [2-3]. We implemented the Arabidopsis multinetwork along with other novel data visualization and analysis techniques we have developed [4] into a common software platform “VirtualPlant v1.0” (www.virtualplant.org) that enables seamless information exploration across diverse data-types and iterative analysis, an essential component of the Systems Biology cycle. VirtualPlant, which currently has a community of 750 registered users from 36 countries including 15 different companies, is the result of a long-standing and highly successful collaboration between biologists and math and computer scientists, notably Gloria Coruzzi (NYU Biology), Dr. Dennis Shasha (NYU Courant), Dr. Manpreet Katari (NYU Biology), and Dr. Rodrigo Gutierrez (Catolica U, Chile).
The ultimate goal of our Plant Systems Biology studies is to predictively model how internal and external perturbations affect processes, pathways, and networks controlling plant growth and development [1]. Succeeding in this endeavor will allow us to (i) explain mechanistically how molecular network changes occur to evoke responses and (ii) predict molecular network states under untested conditions or after modifications. Such predictive network models are useful for developing strategies that intervene in molecular networks for practical applications. To enable such Systems Biology approaches in plants, we developed informatic, visualization, and statistic tools to enable dynamic modeling and visualization of molecular networks operating within cells. The Arabidopsis multinetwork is a first step towards a molecular wiring diagram of the plant cell. This qualitative network model of the Arabidopsis genome contains multiple sources of evidence for all available information on gene, protein, and RNA interactions, including 16,562 nodes and 97,423 interactions [1]. Queries of this qualitative Arabidopsis multinetwork with quantitative transcriptome data result in sub-networks that can be analyzed for network properties, including connectivity, to provide focus for biological hypotheses and testing. Examples of validated hypotheses derived from queries of this multinetwork are described in [2-3]. We implemented the Arabidopsis multinetwork along with other novel data visualization and analysis techniques we have developed [4] into a common software platform “VirtualPlant v1.0” (www.virtualplant.org) that enables seamless information exploration across diverse data-types and iterative analysis, an essential component of the Systems Biology cycle. VirtualPlant, which currently has a community of 750 registered users from 36 countries including 15 different companies, is the result of a long-standing and highly successful collaboration between biologists and math and computer scientists, notably Gloria Coruzzi (NYU Biology), Dr. Dennis Shasha (NYU Courant), Dr. Manpreet Katari (NYU Biology), and Dr. Rodrigo Gutierrez (Catolica U, Chile).
References
[1] Katari et. al. (2010) Plant Physiol. 152:500-15;
[2] Gutiérrez et. al. (2007) Genome Biol. 8:R7;
[3] Gutiérrez et. al. (2008) Proc. Natl Acad Sci USA. 105:4939-4944;
[4] Poultney et. al. Bioinformatics 23:259-61