A functional phylogenomic approach to seed plant evolution
The goal of our work in Evolutionary Genomics is to develop methods that enable us to explore and compare the genomes of diverse plant species to identify genes associated with the important agronomic traits that have evolved in nature. Working as a team of plant systematists, genomicists, and computer scientists, we have developed approaches and tools to enable functional genomic studies within a phylogenetic context. Our goal was to generate and use phylogenomic trees not simply to identify the relationship between species, but more importantly to identify the genes and associated biological processes (e.g. GO terms) that provide positive support for key nodes of trait evolution within plant phylogenies. As proof-of-principle, we conducted a functional phylogenomic analysis of the primitive gymnosperms, the species in which seeds first evolved. We first developed tools to enable genome-scale ortholog identification within a parsimony framework (OrthologID) [1]. We next developed an automated pipeline for the construction of phylogenomic-scale trees, to include all fully sequenced plant genomes and those with > 2,000 unigenes. The resulting matrix and phylogenomic tree, “BigPlantv1.0,” is composed of 22,833 sets of orthologs from the genomes of 150 plant species in 101 genera, analyzed in a combined analysis [2]. This phylogenomic framework enables us to identify overrepresented functional gene categories (GO terms) at major nodes in seed plant phylogeny, revealing critical genes important to angiosperm evolution. This analysis suggests, for example, that RNA interference (RNAi) played a significant role in the divergence of monocots from other angiosperms, a prediction that has experimental support in Arabidopsis and rice. BigPlant v1.0 is now a community resource that enables researchers to explore the genomic origins of plant diversification (http://nypg.bio.nyu.edu/bp/). This project is the result of a highly successful collaboration between PIs at New York University (Gloria Coruzzi), The New York Botanical Garden (Dennis Stevenson), The American Museum of Natural History (Rob DeSalle), and Cold Spring Harbor Labs (Dick McCombie, R. Martienssen).
Most Recently…
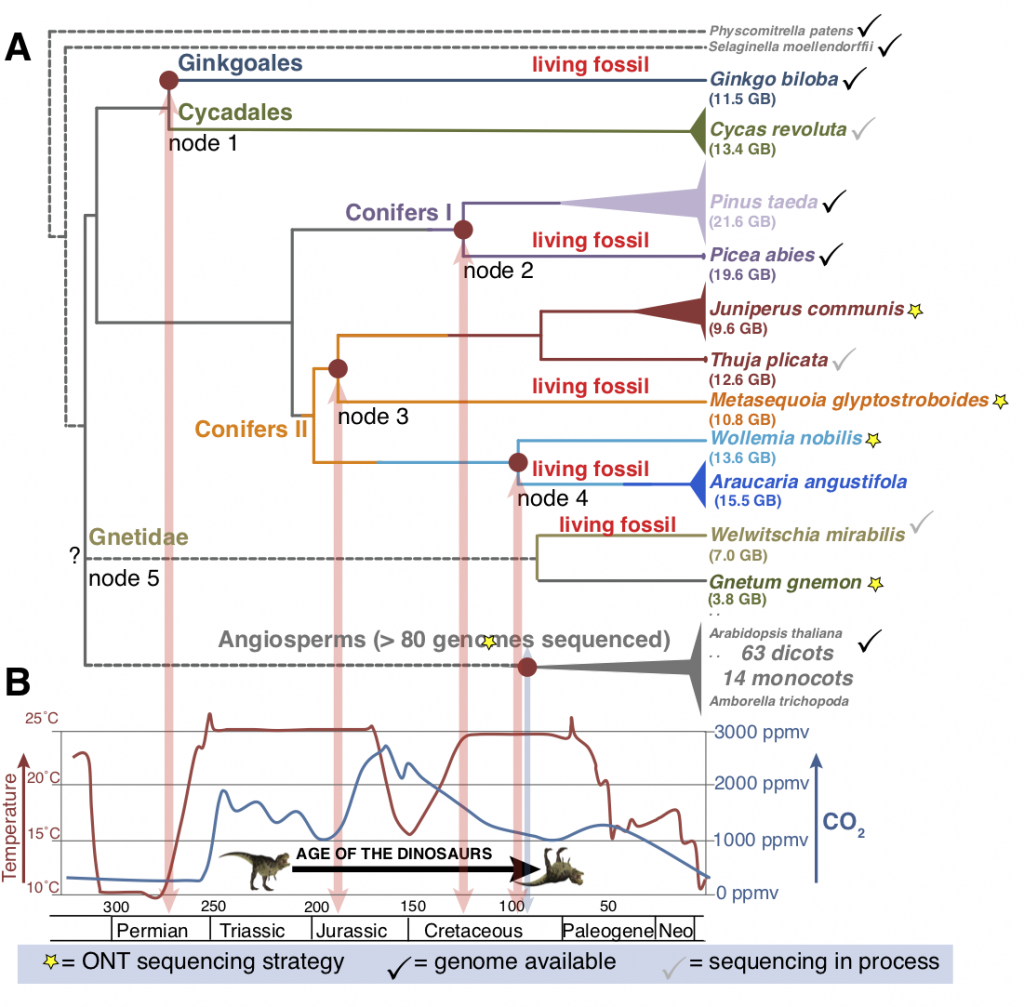
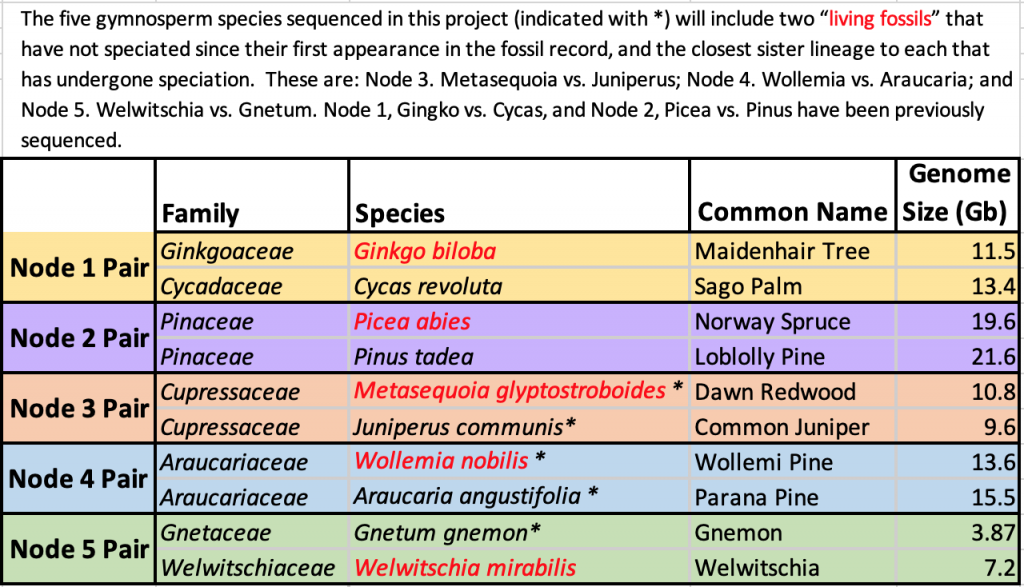
The Living Fossils project is a collaboration of the New York Plant Genomics Consortium (NYBG, AMNH/SUNY, NYU, Purdue, JHU, and CSHL) to sequence and analyze the genomes of 5 gymnosperm species. Living Fossils refers to species that have survived an extensive period of climate change without speciation. These gymnosperms were selected to complement previously sequenced genomes and provide pairs composed of one Living Fossil and their radiated phylogenetic gymnosperm sister pair. These nodes represent unprecedented data from the gymnosperm clade and represent key phylogenetic points for comparison within gymnosperms.
References
[1] Chiu et al (2006) Bioinformatics. 22: 699-707;
[2] Lee et al (2011) PLoS Genetics7(12): e1002411
