November 2025
We are thrilled to announce that our lab’s latest research, “Developmentally regulated genes drive phylogenomic splits in ovule evolution,” has been published in Nature Communications. By combining comparative genomics, developmental biology, and phylogenetic analyses, we reveal that changes in gene regulation can act as a powerful force shaping the diversity of ovules, one of the most critical reproductive structures in plants. Congratulations to the long-standing New York Plant Genomics Consortium for their success and hard work, and our PhD Student Veronica Sondervan! https://www.nature.com/articles/s41467-025-65399-3
June 2025
A new paper published by the Coruzzi Lab titled “Model-to-crop conserved NUE Regulons enhance machine learning predictions of nitrogen use efficiency” appears in a special focus issue of The Plant Cell, “Translational Research from Arabidopsis to Crop Plants and Beyond,” which recognizes the 25th anniversary of the publication of the Arabidopsis genome sequence. DOI: 10.1093/plcell/koaf093. https://academic.oup.com/plcell/article/37/5/koaf093/8129742
Congrats to Ji Huang and all authors on their tremendous work!
August 2024
Key Visions: A tribute to Joe L. Key
Founder of plant molecular biology and its community
I first became introduced to the nascent plant molecular biology community at the 2nd Plant Molecular Biology Gordon conference, chaired by Joe Key in 1981. That was a seminal meeting in our field, and my connections to Joe Key and his colleagues grew over the years.
This tribute to Joe Key is for his vision and leadership at the frontier of plant molecular biology, both scientifically and for community building. He created the plant molecular field that we all so richly cherish. We are truly grateful for his vision and mourn his passing on Aug. 19, 2024.
August 2023
NEW! preprint of the “Living Fossil” Wollemia nobilis genome available on BioRxiv
Featured Press Releases for Wollemi Pine Article!
Mystery of ‘living fossil’ tree frozen in time for 66 million years finally solved
Ancient Wollemi Pine: A Living Fossil Threatened with Extinction in Australia
“Living Fossil” Wollemi Pine Is Ancient, Inbred, And In A Lot Of Trouble
Solved: The Enigma of the ‘Living Fossil’ Tree Preserved for 66 Million Years
November 2022
Frontiers article written by Coruzzi Lab Post Doc Carly Shanks!
Validation of a high-confidence regulatory network for gene-to-NUE phenotype in field-grown rice
Front. Plant Sci., 25 November 2022
Sec. Plant Physiology
https://doi.org/10.3389/fpls.2022.1006044
October 2022
The TARGET System: Rapid Identification of Direct Targets of Transcription Factors by Gene Regulation in Plant Cells
Our TARGET system is published in as part of the Methods in Molecular Biology book series on Transcription Factor Regulatory Networks. (MIMB,volume 2594)
Transcription Factor Regulatory Networks pp 1–12 Matthew D. Brooks, Kelsey M. Reed, Gabriel Krouk, Gloria M. Coruzzi & Bastiaan O. R. Bargmann Protocol First Online:
DOI: 10.1007/978-1-0716-2815-7_1
December 2021
Gloria Coruzzi is one of 24 NYU professors named the world’s most-cited researchers! Read about this exciting news in a recent article in “Washington Square News”, NYU’s Independent Student Newspaper.

Our recent paper, “Plant ecological genomics at the limits of life in the Atacama Desert” has been cited by PNAS, to be in the top 5% of their Altmetric Attention Score. This score provides an indicator of the amount of attention that a PNAS article has received. So far, our paper has been written about in 37 news stories from 30 outlets!
November 2021
NEW PNAS paper published on research led by collaborator Dr. Rodrigo A. Gutierrez, in Chile. Thank you for letting us join you on this 10- year journey!
Plant ecological genomics at the limits of life in the Atacama Desert
Featured Press Releases for PNAS Article!
Genetic Engineering & Biotechnology News
October 2021
NSF’s Research News article covering NSF PGRP NutriNet Machine Learning paper:
Machine learning uncovers ‘genes of importance’ in agriculture
https://www.nsf.gov/discoveries/disc_summ.jsp?cntn_id=303673
September 2021
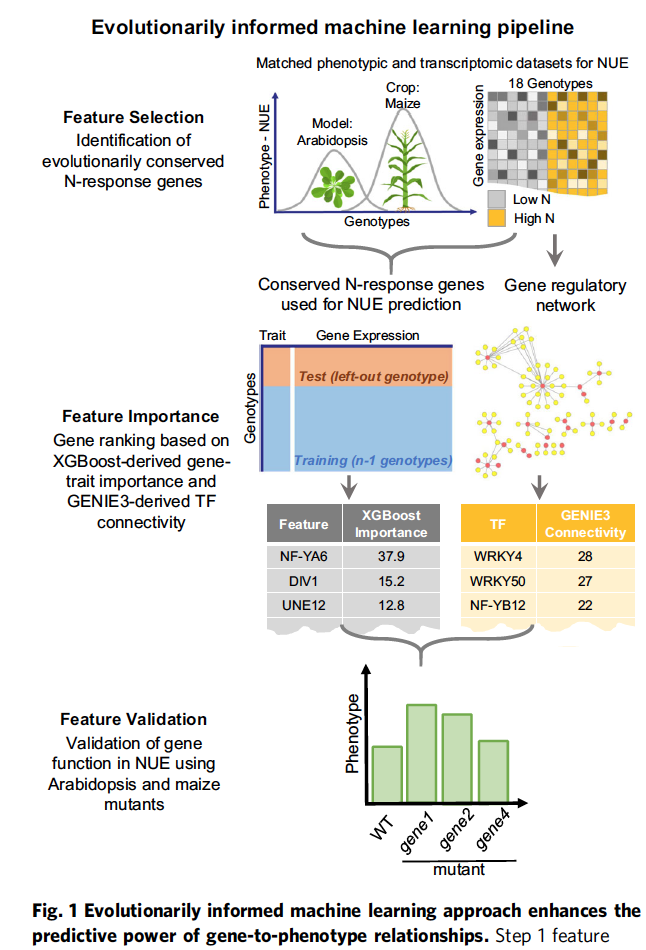
Nature Communications paper titled “Evolutionarily informed machine learning enhances the power of predictive gene-to-phenotype relationships.” published September 2021.
Click here to read the article on Nature Communications!
Click here to read the wonderful NYU Press Release!
May 2020
Congratulation to all authors!
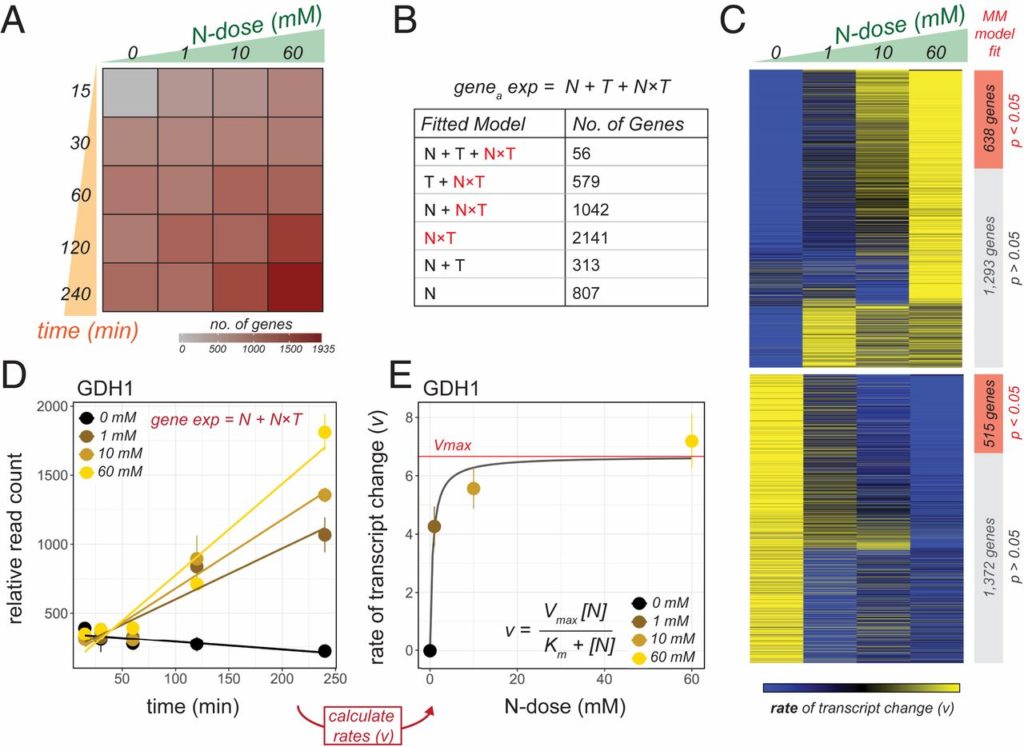
Happy to share our newest story “Nutrient dose-responsive transcriptome changes driven by Michaelis–Menten kinetics underlie plant growth rates” on PNAS!
March 2020
Congratulations to Alvarez and Schinke et al.
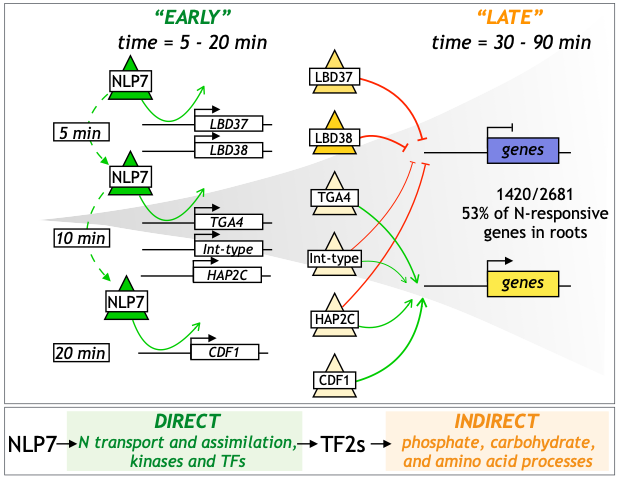
NLP7 paper recommendation in F1000Prime.
Press release on NLP7
Link to AAAS
Link to NYU Press
Link to Manuscript
April 2019
Gloria Coruzzi is elected into the National Academy of Sciences

Link to NYU Press Release
Link to NAS Press Release
Link to NYU Tweet

May 2018
Biologists identify temporal logic of regulatory genes affecting nitrogen use efficiency in plants
Discovery has implications for global food production, sustainable agriculture
Kranthi Varala, Amy Marshall-Colón, Jacopo Cirrone, Matthew D Brooks, Sophie Léran, Shipra Mittal, Angelo V Pasquino, Tara M Rock, Molly B Edwards, Grace J Kim, W Richard McCombie, Dennis Shasha & Gloria M Coruzzi (2018). “The transcriptional logic of dynamic regulatory networks that underlie nitrogen signaling and use in plants.” Proc Natl Acad Sci U S A. 2018 May 16. pii: 201721487. doi: 10.1073/pnas.1721487115. [Epub ahead of print]
PMID: 29769331
The transcriptional Logic of Nitrogen Regulatory Networks.
- https://www.sciencedaily.com/releases/2018/05/180514151918.htm
- https://phys.org/news/2018-05-biologists-id-temporal-logic-regulatory.html
- https://sciencenews.newspedia.online/2018/05/15/biologists-id-temporal-logic-of-regulatory-genes-affecting-nitrogen-use-efficiency-in-plants/
- http://news.agropages.com/News/NewsDetail—26318.htm
- http://www.isaaa.org/kc/cropbiotechupdate/article/default.asp?ID=16443
- http://www.canada-goosejackets.com.co/?p=62023
- http://www.purdue.edu/newsroom/releases/2018/Q2/method-for-identifying-key-regulator-genes-may-speed-improvements-in-fertilizer-use-and-other-efficiencies.html
- http://www.findclimateanswers.com/biologists-id-temporal-logic-of-regulatory-genes-affecting-nitrogen-use-efficiency-in-plants/
- https://twitter.com/howplantswork
- https://www.researchgate.net/publication/325183037_Temporal_transcriptional_logic_of_dynamic_regulatory_networks_underlying_nitrogen_signaling_and_use_in_plants
- https://www.scoop.it
September 2017
NSF Science Nation video released on Coruzzi lab research: Genomic science uncovers genes that enable plants to grow more with less fertilizer. Click here to see the video.
August 2017

In memory and tribute to Alexandra Page Clark: Student, Research Scientist, Friend
April 2017

Find the article here
June 2016

They are (from left to right): XaoFei Lin, Kristen Lee, Apurva Parikh (back), Tanim Jain, Mohammad Sadic, Priyanka Srivastava, and John Yuen.
In Atlas of Science:
From milliseconds to lifetimes: dynamic behavior of transcription factors in gene networks
Link to Summary: http://atlasofscience.org/from-milliseconds-to-lifetimes/
Conserved nitrogen-regulated network modules in rice
Link to Summary: http://atlasofscience.org/conserved-nitrogen-regulated/
In Bioessays:
“Hit-and-run”: Transcription factors get caught in the act
Link to Article: http://onlinelibrary.wiley.com/doi/10.1002/bies.201400186/epdf
R Workshop at NYU CGSB
July 20th – 24th, 2015
Manny Katari teaches an R Course at NYU’s Center for Genomics and Systems Biology. Students were Postdoctoral Fellows, Graduate Students, and Undergraduate Students.

The syllabus included training modules on R Data Structures, Importing and Exporting Data in R, Using the Apply Function, Basic Plot Functionality, and Advanced Plots.
June 23rd, 2014
The Coruzzi Lab discovers a new “hit-and-run” model of transcription
Dr. Coruzzi led a team of genome scientists that has identified a “hit-and-run” mechanism that allows regulatory proteins in the nucleus to adopt a “Tom Sawyer” behavior when it comes to the work of initiating gene activation.
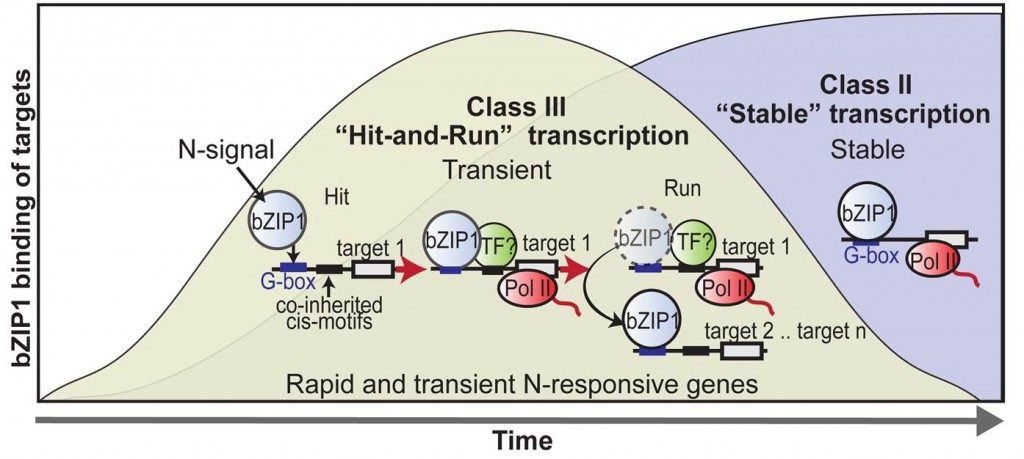
Their research, which appears in the Proceedings of the National Academy of Sciences, focuses on transcription factors—proteins that orchestrate the flow of genetic information from DNA to messenger RNA (mRNA). Their results show how transcription factors (TFs) activate mRNA synthesis of a gene, and leave the scene – in a model termed “hit-and-run” transcription.
“Much like Mark Twain’s Tom Sawyer who begins to paint Aunt Polly’s fence, and then convinces others that they are privileged to join in before leaving to relax, this pioneer transcription factor binds to a gene promoter to initiate transcription and then leaves, recruiting its friends to continue work it started,” explains New York University Biology Professor Coruzzi, the study’s senior author.
The transcription factor under study is crucial to activating genes needed to respond to nitrogen, a nutrient signal that is the rate-limiting element in plant growth.
This work was supported by grants from the National Institutes of Health (NIH) (R01-GM032877, R01-GM078270), an NIH National Research Service Award (GM095273), the National Science Foundation (MCB-0929338, DBI-0923128), France’s National Research Agency and National Center for Scientific Research.
Alessia Para, Ying Li, Amy Marshall-Colón, Kranthi Varala, Nancy J. Francoeur, Tara M. Moran, Molly B. Edwards, Christopher Hackley, Bastiaan O. R. Bargmann, Kenneth D. Birnbaum, W. Richard McCombie, Gabriel Krouk, and Gloria M. Coruzzi. (2014) “Hit-and-run transcriptional control by bZIP1 mediates rapid nutrient signaling in Arabidopsis.” Proceedings of the National Academy of Sciences. doi:10.1073/pnas.1404657111
December 2013
Coruzzi Lab Celebrates the Holidays
The Coruzzi Lab celebrates the holiday season at the Washington Square Park tree lighting ceremony.

September 11, 2013
A Walk Through Gloria Coruzzi’s Plant Genome Wonderland
An article and slideshow on the work of Dr. Gloria Coruzzi and her lab at New York University’s Center for Genomics and Systems Biology is currently being featured on the NYU Stories website.

Dr. Coruzzi led a tour of NYU’s state-of-the-art Center for Genomics and Systems Biology to showcase the ongoing projects in her lab. She explained how access to such a facility has radically changed the way she approaches research. “Here at NYU, before we had the Center, we had started doing genome-scale experiments but we didn’t have the wide range of high-throughput genomic capabilities that we have now in our Core facility,” Coruzzi explains. “The openness between the labs and the bioinformatics [suite] has just exploded our ability to do genome science.”
March 19, 2012
NYC-Area High School Students Working with NYU Biologists Named Intel Science Talent Search Finalists
The Intel STS is the nation’s most prestigious pre-college science competition. This year, Intel STS chose its 40 finalists from 1,839 applicants from around the U.S. Previous Intel STS winners have gone on to capture more than 100 of the world’s most distinguished science and math honors, including seven Nobel Prizes and four National Medals of Science.

Angela Fan, finalist for Intel STS, worked with CGSB researchers. Fan, a student at Stuyvesant High School, studied root nutrient foraging in the laboratory of Professor Gloria Coruzzi. She applied a morphometric approach to quantifying the developmental plasticity space of different ecotypes of the model plant species Arabidopsis thaliana in laboratory and natural environments.
January 18, 2012
Four NYC-Area High School Students Working With NYU Biologists Name Intel Science Fair Semifinalists

Four high school students working on science-fair research projects with scientists at NYU’s Center for Genomics and Systems Biology (CGSB) are among the 300 national semifinalists for the international INTEL Science Talent Search competition for 2012. These high school students worked on a range of projects, including plant nutrient dynamics, protein structure prediction, nematode genomics, and bacterial spore development.
Angela Fan (Stuyvesant High School, NYC), who worked in the plant genomics and systems biology laboratory of Professor Gloria Coruzzi, studied root nutrient foraging. She applied a morphometric approach to quantifying the developmental plasticity space of different ecotypes of the model plant species Arabidopsis thaliana in laboratory and natural environments.
December 15, 2011
Scientists Create Largest-Ever Genome Tree of Life for Seed Plants
Scientists at New York University’s Center for Genomics and Systems Biology, the American Museum of Natural History, Cold Spring Harbor Laboratory, and the New York Botanical Garden have created the largest genome-based tree of life for seed plants to date. Their findings, published today in the journal PLoS Genetics, plot the evolutionary relationships of 150 different species of plants based on advanced genome-wide analysis of gene structure and function. This new approach, called “functional phylogenomics,” allows scientists to reconstruct the pattern of events that led to the vast number of plant species and could help identify genes used to improve seed quality for agriculture.

“Ever since Darwin first described the ‘abominable mystery’ behind the rapid explosion of flowering plants in the fossil record, evolutionary biologists have been trying to understand the genetic and genomic basis of the astounding diversity of plant species,” said Rob DeSalle, a corresponding author on the paper and a curator in the Museum’s Division of Invertebrate Zoology who conducts research at the Sackler Institute for Comparative Genomics. “Having the architecture of this plant tree of life allows us to start to decipher some of the interesting aspects of evolutionary innovations that have occurred in this group.”
