EVONET:
A Phylogenomic and Systems Biology approach to identify genes underlying plant survival
in marginal, low-Nitrogen soils.
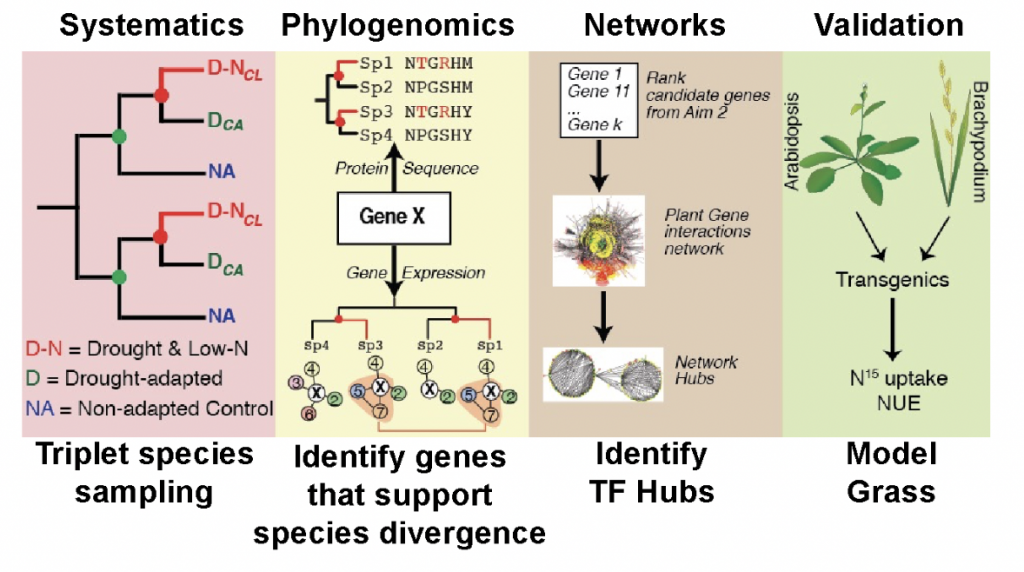
This DOE BER sustainability project aims to identify the key genes and gene
regulatory networks that enable “extreme survivor” plants to adapt and grow in marginal, nitrogen
(N)-poor soils in the hyper-arid Atacama Desert in Chile. These “extreme survivor” species cover
the main branches in flowering plants and include 7 grass species of particular interest for biofuels.
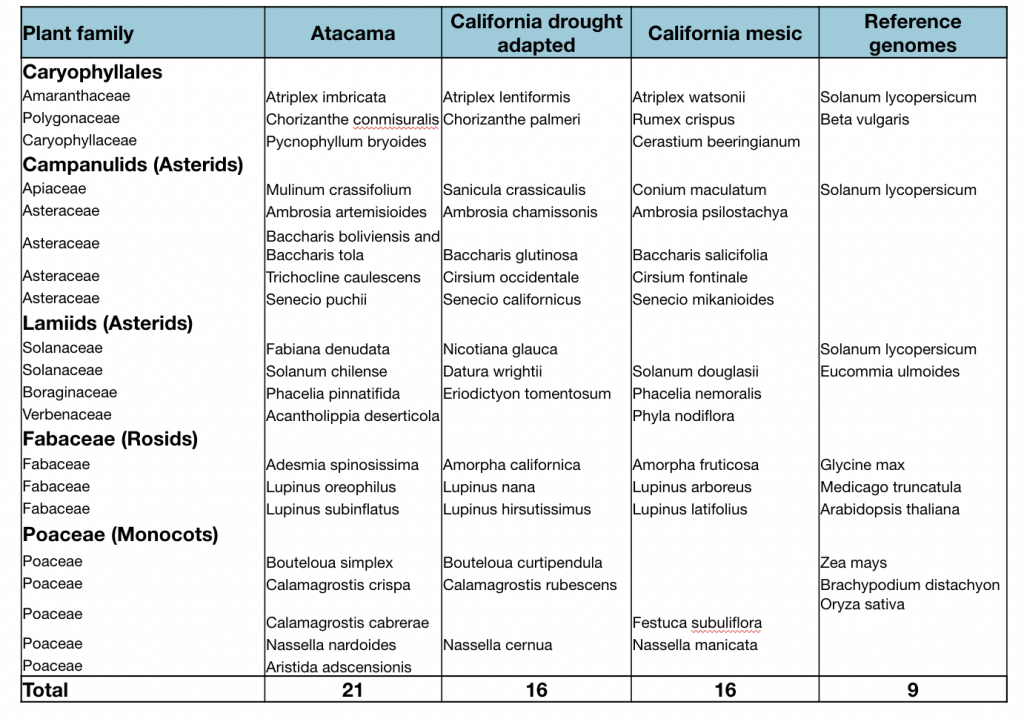
We focus on 28 “extreme survivor” Atacama species and compare their encoded genes to
Californian “sister” species that live in a N-replete conditions in arid (27 species) or mesic (27
species) environments. Deep RNA-sequencing of these “triplet species” was used to fuel a
phylogenomic analysis that can identify genes that support the evolutionary divergence of the
extreme survivors in Atacama Desert from their sister species in California. The genes thus
identified will help to discover the mechanisms underlying physiological and developmental
processes that allow plant survival in nitrogen-poor, dry soils. The genes and network modules so
uncovered can potentially be translated to biofuel crops to greatly increase biomass and nitrogen
use efficiency in marginal, low-fertility soils.



Most recent Publication:

Plant ecological genomics at the limits of life in the Atacama Desert
The Atacama Desert in Chile—hyperarid and with high–ultraviolet irradiance levels—is one of the harshest environments on Earth. Yet, dozens of species grow there, including Atacama-endemic plants. Herein, we establish the Talabre–Lejía transect (TLT) in the Atacama as an unparalleled natural laboratory to study plant adaptation to extreme environmental conditions. We characterized climate, soil, plant, and soil–microbe diversity at 22 sites (every 100 m of altitude) along the TLT over a 10-y period. We quantified drought, nutrient deficiencies, large diurnal temperature oscillations, and pH gradients that define three distinct vegetational belts along the altitudinal cline. We deep-sequenced transcriptomes of 32 dominant plant species spanning the major plant clades, and assessed soil microbes by metabarcoding sequencing. The top-expressed genes in the 32 Atacama species are enriched in stress responses, metabolism, and energy production. Moreover, their root-associated soils are enriched in growth-promoting bacteria, including nitrogen fixers. To identify genes associated with plant adaptation to harsh environments, we compared 32 Atacama species with the 32 closest sequenced species, comprising 70 taxa and 1,686,950 proteins. To perform phylogenomic reconstruction, we concatenated 15,972 ortholog groups into a supermatrix of 8,599,764 amino acids. Using two codon-based methods, we identified 265 candidate positively selected genes (PSGs) in the Atacama plants, 64% of which are located in Pfam domains, supporting their functional relevance. For 59/184 PSGs with an Arabidopsis ortholog, we uncovered functional evidence linking them to plant resilience. As some Atacama plants are closely related to staple crops, these candidate PSGs are a “genetic goldmine” to engineer crop resilience to face climate change.
Gil Eshel, Viviana Araus, Soledad Undurraga, and Rodrigo A. Gutiérrez
November 1, 2021118 (46) e2101177118https://doi.org/10.1073/pnas.2101177118